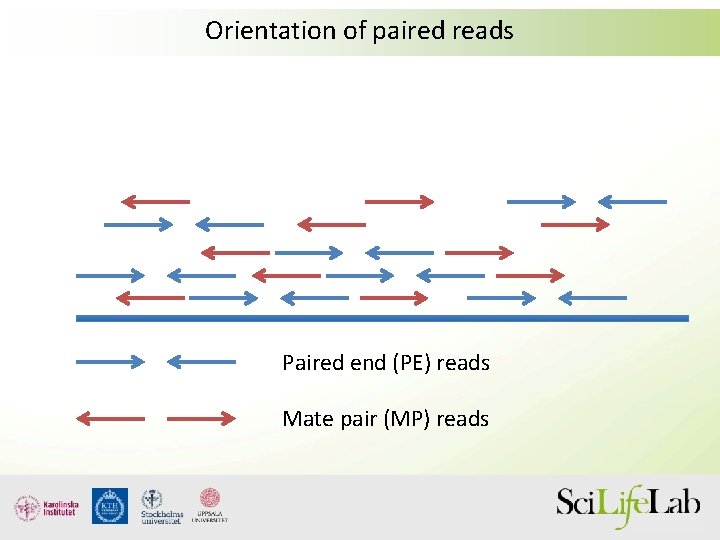
paired end sequencing vs mate pair
In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.

Mate Pair Made Possible Enseqlopedia
Introduction to Mate Pair Sequencing.

. The insert size on classic paired-end is smaller about 500bp. PacBio SMRT 2017 2018 From PacBio web site. Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported two files R1 and R2 often refereed to as mates files R1first mates R2second mates.
Mate pair vs paired end. These fragments are. As with everything you get what you pay for- paired end sequencing will always be the better option but for differential expression analysis it is likely not worth the additional cost.
Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. For example if you have a 300bp contiguous fragment the machine will sequence eg. 1 shows a schematic view of an Illumina paired-end read.
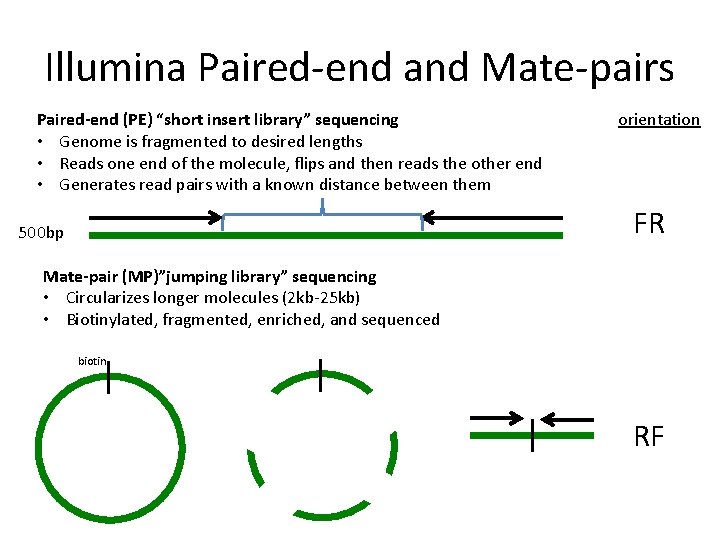
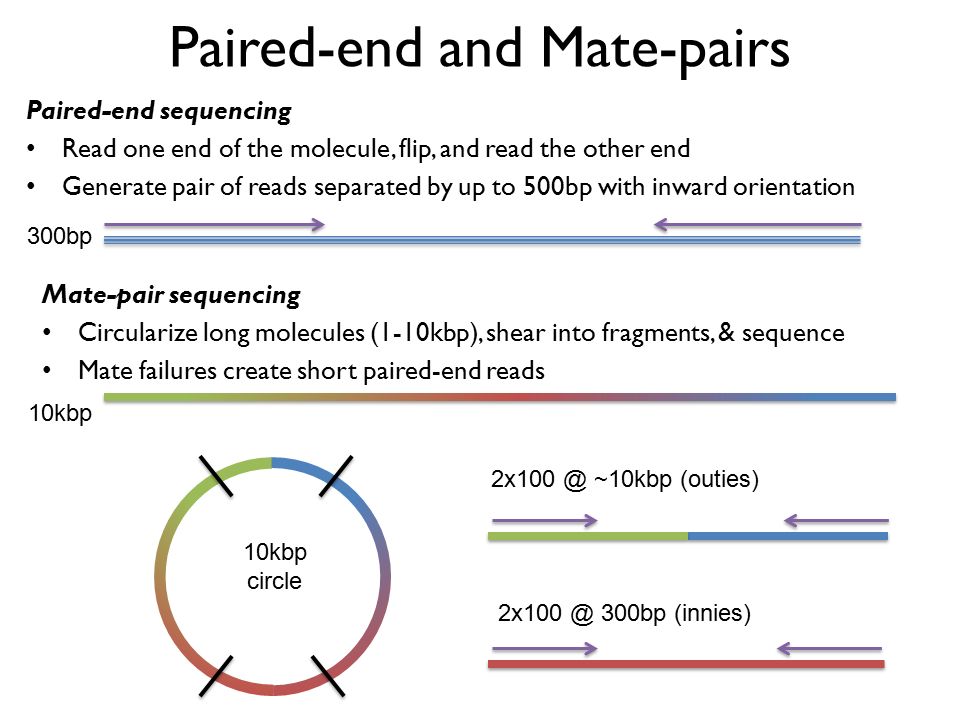
The mate-pair data allows you to do things like assemble across repetitive regions that you cannot resolve with paired end data. Scaffolds NNN Contigs Illumina Reads Assembly Strategy Paired-end reads Mate-pair reads NNNN Contiging Scaffolding. In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably.
This is known as an FR read forwardreverse in that order. What is adapter sequence. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
Using a combination of short and long insert sizes with paired-end sequencing results in maximal coverage. Introduction to Mate Pair Sequencing. Paired-end tags PET sometimes Paired-End diTags or simply ditags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they theoretically exist together only once in a genome therefore making the sequence of the DNA in between them available upon search if full-genome sequence data is available or upon further.
The adapter sequence is the sequence of the adapter to be trimmed. This can be very helpful e. Methods A novel algorithm that utilizes a suffix array has been specifically designed to compute the uniqueness of paired reads with fixed or variable mate-pair distance.
10-20 kb Long-read Sequencing Platform. Paired-end reading improves the ability to identify the relative positions of various reads in the genome making it much more effective than single-end reading in resolving structural rearrangements. We obtain theoretical read length lower bounds for re-sequencing that are also applicable to paired-end de novo assembly.
While the insert size of mate-pair is. While the underlying principles between PE and MP reads have strong similarities there are inherent differences that are crucial to understand. There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter.
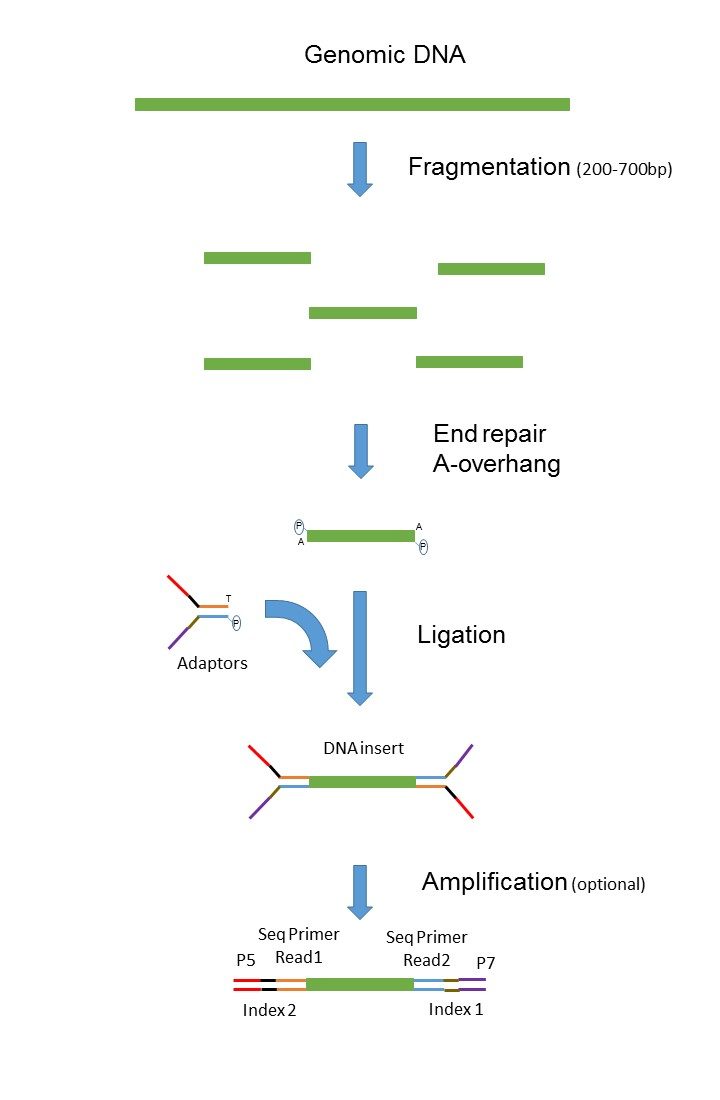
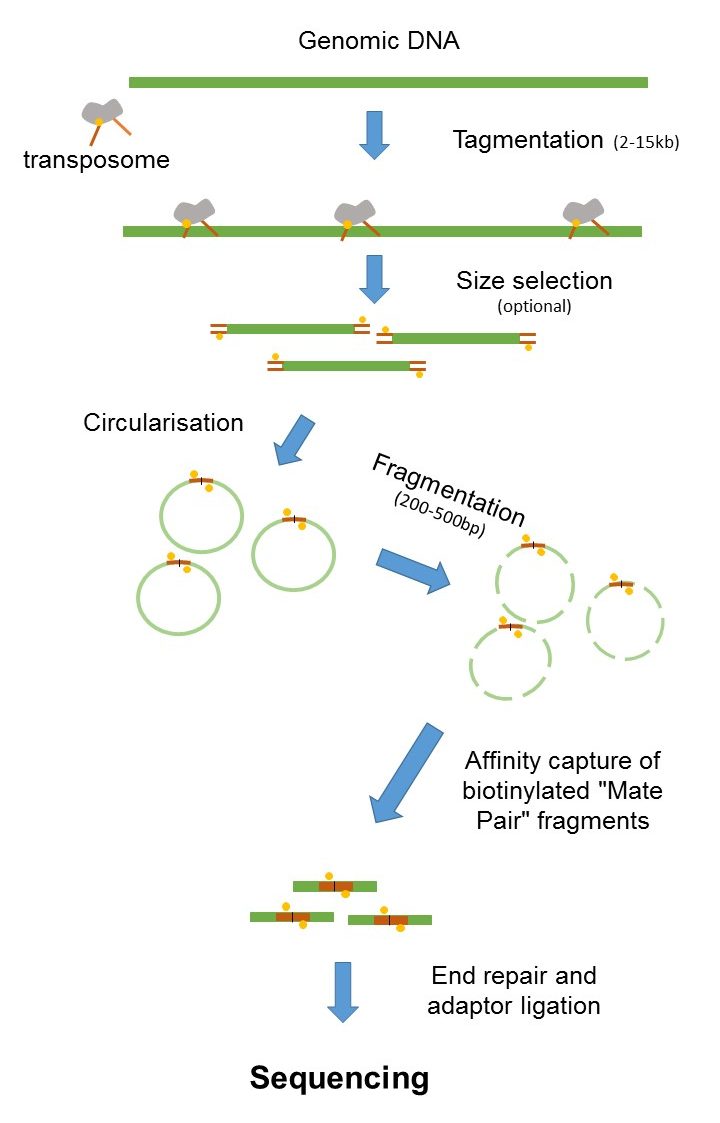
Shortinsert pairedend reads SIPERs and long-insert paired-end reads LIPERs. Paired-end or mate-pair. The figure shows the workflow for mate-pair library preparation for Illumina sequencing.
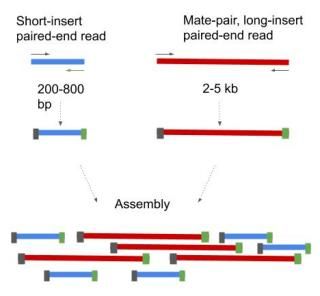
Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. For your De novo genome assembly Fig. Mate pair reads Fragment.
Illumina Paired End Sequencing. This is all for conventional paired-end sequencing. For me mate-pair and classic paired-end are both paired-end reads with the difference that.
Since paired-end reads are more likely to align to a reference the. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first generation sequencing. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. 2 For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. I wondered what the essential differences were between paired-end mate-pair and long read.
I writing here because I have some questions for you. One of the advantages of paired end sequencing over single end is that it doubles the amount of data. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal sequencing coverage across the genome.
Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. What is paired-end and mate pair sequencing.
2 For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. Why is paired end sequencing better.
When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one so that they are pointed towards one another. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment.
Another supposed advantage is that it leads to more accurate reads because if say Read 1 see picture below maps to two different regions of the genome Read 2 can be used to help determine which one of the two regions makes more sense. 1 shows a schematic view of an Illumina paired-end read. Mate pair sequencing is used for various applications applications including.
Since the beginning of 2013 this preparation has been based on Nextera.
Paired End Sequencing France Genomique

Principles For Construction Of Mate Pair Sequencing Libraries A Download Scientific Diagram

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram
De Novo Genome Assembly Using Next Generation Sequence
Introduction To Genome Assembly Tutorial

What Is Mate Pair Sequencing For

What Is Mate Pair Sequencing For
Whole Genome Assembly With Iplant Ppt Video Online Download

Ion Torrent Mate Paired Library Preparation Protocol And Sequencing Download Scientific Diagram

Construction Of Mate Pair Libraries Download Scientific Diagram
De Novo Genome Assembly Introduction Henrik Lantz Nbissci
Why Mate Paired End Paired End And Single End Reads Library To Be Combined For Assembling

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar
Mate Pair Sequencing France Genomique
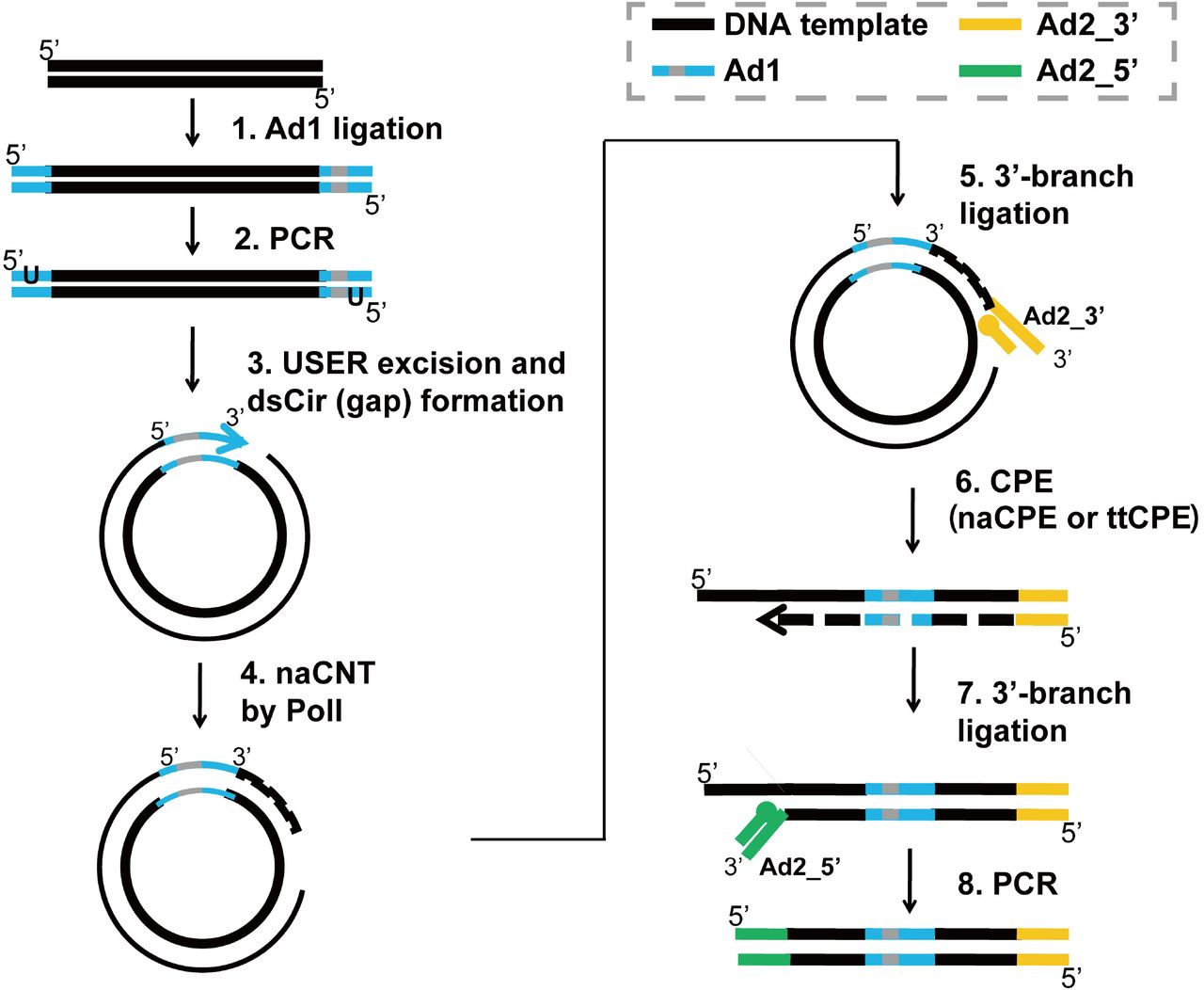
Development Of Coupling Controlled Polymerizations By Adapter Ligation In Mate Pair Sequencing For Detection Of Various Genomic Variants In One Single Assay Biorxiv

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download Scientific Diagram

Principles Of Mate Pair Libraries Construction And The Bioinformatics Download Scientific Diagram
